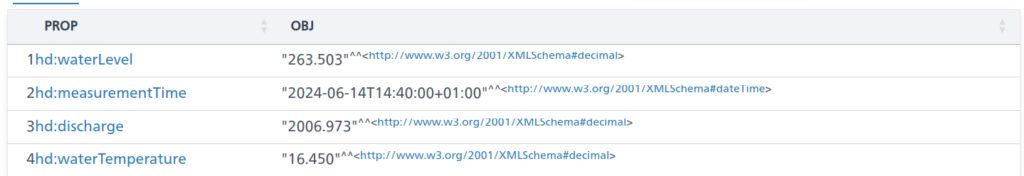
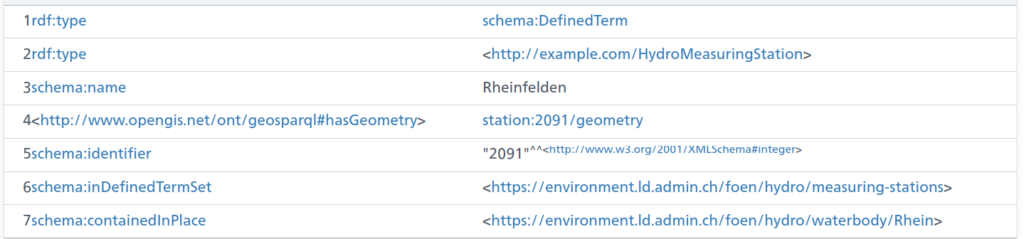
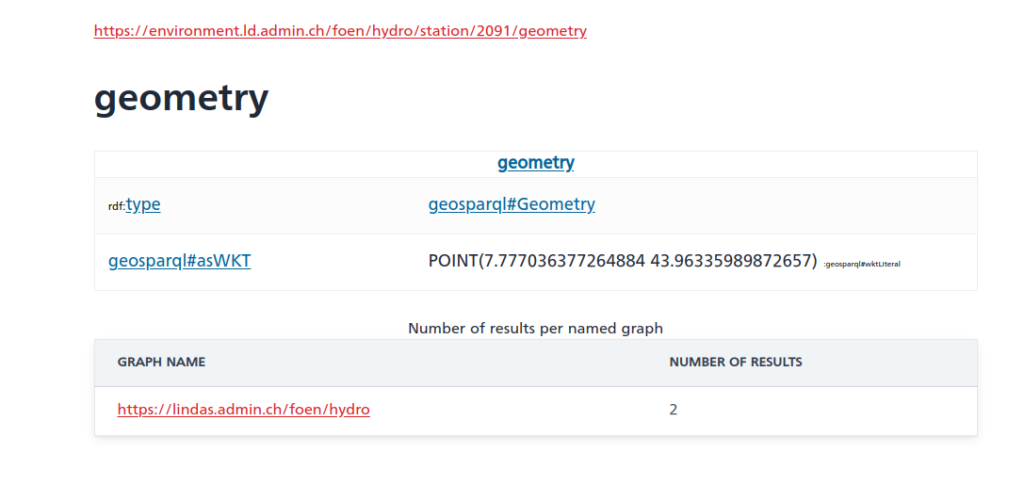
TL;DR;
Make a map of the original and/or fixed coordinates of the hydrological stations of the BAFU using the data from Lindas

The above map is an illustration of the data we can extract with use of a SPARQL query and lindas of the hydrological data published on it. As you can see, there are two sets of points, one in Switzerland (corrected version) and one in the Mediterranean based on the “raw” lindas data (400km offset for some reason).
In my last post, I wanted to see how to get the current data of waterbodies from lindas.admin.ch in one table including geometry information. Now, we of course want to use Python to investigate the data; additionally, we want to extend our existing hydrodata SPARQL query to handle also a subset of stations instead of all stations.
import datetime
import requests
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# The prefixes, mainly to have them seperate
PREFIXES = """
PREFIX schema: <http://schema.org/>
PREFIX hg: <http://rdf.histograph.io/>
PREFIX la: <https://linked.art/ns/terms/>
PREFIX ex: <http://example.org/>
# hydro related prefixes
PREFIX h: <https://environment.ld.admin.ch/foen/hydro/>
PREFIX hd: <https://environment.ld.admin.ch/foen/hydro/dimension/>
PREFIX hgs: <https://environment.ld.admin.ch/foen/hydro/station/>
PREFIX river: <https://environment.ld.admin.ch/foen/hydro/river/observation/>
PREFIX lake: <https://environment.ld.admin.ch/foen/hydro/lake/observation/>
# geo related prefixes
PREFIX wkt: <http://www.opengis.net/ont/geosparql#asWKT>
# hydrodata related prefixes (fake) that we use to reference to the historical data. haha.
PREFIX hydrodata: <https://www.hydrodaten.admin.ch/plots/>
"""
## a function that makes the response to a dataframe, tailored to the hydrodata
# and that also contains a small validation step to remove "outdated" values
def _response2dataframe(response,
validate=True,
time_threshold=datetime.timedelta(minutes=30)):
# return pd.read_csv(io.StringIO(resp.text))
data = response.text.strip().split('\n')
columns = data[0].split(',')
data = [i.strip().split(',') for i in data[1:]]
index = [i[0] for i in data]
ix = columns.index('data')
values = [[i[0]] + i[ix].split(';') for i in data]
data = [[j for _, j in enumerate(i) if _ != ix]
for i in data]
# the dataframe holding all non measurement values
columns.pop(ix)
data1 = pd.DataFrame(columns=columns,
data=data,
index=index)
for col in data1.columns:
data1[col] = pd.to_numeric(data1[col], errors='ignore')
def _list2dict(datalist):
return {i: j for i, j in zip(datalist[::2], datalist[1::2])}
values = {i[0]: _list2dict(i[1:])
for i in values}
ix = values.keys()
columns = []
for i in values.items():
vartypes = list(i[1].keys())
for vartype in vartypes:
if vartype in columns:
pass
else:
columns.append(vartype)
data = [[] for i in columns]
for i in ix:
for colno, colname in enumerate(columns):
value = values[i][colname] if colname in values[i].keys() else ''
data[colno].append(value)
data = np.transpose(data)
data2 = pd.DataFrame(columns=columns, data=data, index=ix)
for col in data2.columns:
if col != 'measurementTime':
data2[col] = pd.to_numeric(data2[col], errors='coerce')
data = pd.concat([data1, data2], axis=1)
data['query_time'] = pd.to_datetime(data['query_time'])
data['measurementTime'] = pd.to_datetime(data['measurementTime'])
if validate:
time_threshold = pd.to_timedelta(time_threshold)
good = data['query_time'] <= data['measurementTime'] + time_threshold
# for illustration
bad = data['query_time'] >= data['measurementTime'] + time_threshold
data['measurementAge'] = data['query_time'] - data['measurementTime']
data = data[good]
return data
def get_hydrodata(station_ids=None,
return_as_dataframe=True,
validate=True,
time_threshold=datetime.timedelta(minutes=30)):
query = [
"""# what we want to get out of the query as "nicely" formatted data
SELECT ?station_id ?station_name ?station_type ?station_type_nice
?latitude ?longitude ?station_iri ?station_data_iri ?station_geometry_iri
(group_concat(?measurement_type_measurement_value;SEPARATOR=";") as ?data)
?query_time
# add the https://hydrodaten.admin.ch url for the data that go back 7 days (actually 40 days sometimes, see dev console)
(if(contains(?data,"discharge"), uri(concat(str(hydrodata:), 'p_q_7days/', ?station_id, '_p_q_7days_de.json')), "")
as ?hydrodata_url_discharge_waterlevel)
(if(contains(?data,"temp"), uri(concat(str(hydrodata:), 'temperature_7days/', ?station_id, '_temperature_7days_de.json')), "")
as ?hydrodata_url_temperature)
"""
# where part including initial setup of measurement types
"""
WHERE { ?station ?prop ?value.
filter( ?prop in (ex:isLiter, hd:measurementTime, hd:waterTemperature, hd:waterLevel, hd:discharge))
""",
# generating data and variables part
"""
bind(replace(replace(
str(?station), str(river:), "", "i"),
str(lake:), "", "i")
as ?station_id)
# the above is equivalent to this in theory but the above uses prefixes so would be easier to change
# in case of changes
# bind(strafter(str(?station), "observation/") as ?station_id2)
# get the kind of station this is (whether lake or river) - the hierarchy/ontology here is weird as all are measurement stations
# or observation but the distinction is river/lake first then observation
bind(strbefore(strafter(str(?station), str(h:)), "/") as ?station_type)
# uppercase the first letter -> why is there no capitalize function in SPARQL :throw_table:
bind(concat(ucase(substr(?station_type,1,1)), substr(?station_type,2,strlen(?station_type)-1)) as ?station_type_nice)
# remove some filler words from the data
bind(replace(str(?prop), str(hd:), "", "i") as ?measurement_type)
# convert the actual measurement value to a literal
bind(str(?value) as ?measurement_value)
# introduce the time of the query to (maybe) see if the values are old
bind( str(now()) as ?query_time)
# combine the type of measurement with the value so we can group by and not loose information
bind(concat(?measurement_type, ';', ?measurement_value, '') as ?measurement_type_measurement_value)
# start making IRIs so we can check/link back to lindas
# the IRI containing information of the station itself (not linked to river/lake)
bind(IRI(concat(str(hgs:), ?station_id)) as ?station_iri)
# generate the geometry IRI
bind(IRI(concat(str(hgs:), concat(?station_id,"/geometry"))) as ?station_geometry_iri)
# generate the station IRI that holds the data information
bind(IRI(concat(str(h:), ?station_type, "/observation/", ?station_id)) as ?station_data_iri)
# get the information about the location of the station
?station_geometry_iri wkt: ?coordinates.
# also get the name
?station_iri schema:name ?stat_name.
bind(replace(?stat_name, ',', ';') as ?station_name)
# simplifiy the geometry to lon/lat in array form
BIND(STRBEFORE(STRAFTER(STR(?coordinates), " "), ")") AS ?latitude)
BIND(STRBEFORE(STRAFTER(STR(?coordinates), "("), " ") AS ?longitude)
}""",
# grouping part
"""
group by # the station information
?station_id ?station_type ?station_type_nice ?station_name
?latitude ?longitude
# the IRIs
?station_iri ?station_data_iri ?station_geometry_iri
# validation information
?query_time
""",
# sorting part
"""
order by ?station_id
"""
]
if station_ids is not None:
# per default, get all stations but if some stations are given uses
# this modication instead
station_ids = station_ids if isinstance(
station_ids, list) else list(station_ids)
station_ids = '"' + '","'.join([str(i) for i in station_ids]) + '"'
query.insert(1, f'filter (?station_id in ({station_ids}))')
query = PREFIXES + '\n'.join(query)
resp = requests.post("https://ld.admin.ch/query",
data="query=" + query,
headers={
"Content-Type": "application/x-www-form-urlencoded; charset=utf-8",
"Accept": "text/csv"
}
)
resp.encoding = "utf-8"
if return_as_dataframe:
resp = _response2dataframe(resp,
validate=validate,
time_threshold=time_threshold)
return resp, query
if __name__ == '__main__':
df, query = get_hydrodata()
subset = [2091, 2613]
df_subset, query_subset = get_hydrodata(station_ids=subset,
return_as_dataframe=True)
A first approach is given by the lindas help which showcases the use of the typical request together with pandas. The example is straightforward but not usually how we’ll have data (more than 3 columns ..) that are useful with pandas. As we’ve seen in the last exploration of lindas, we can write a query that gives us a row for each station of the hydrological network of the BAFU. To be consistent with the example, we want a CSV return, which means replacing all commas with something better (especially the names of stations sometimes do have a comma inside). Other than that, the query that we used before also works directly via Python and we only extended it with the option to additionally filter with station_ids by adding the below (which adds a very simple filter into the query):
if station_ids is not None:
# per default, get all stations but if some stations are given uses
# this modication instead
station_ids = station_ids if isinstance(
station_ids, list) else list(station_ids)
station_ids = '"' + '","'.join([str(i) for i in station_ids]) + '"'
query.insert(1, f'filter (?station_id in ({station_ids}))')
Additionally, as I didn’t manage to split the values again into separate columns after the grouping (or only with a lot of needless wordiness), I wrote a small converter function that mainly extracts the general information (meta-information if you will like latitude/longitude or similar) and converts the actual measurement data into columns and concatenates the two.
## a function that makes the response to a dataframe, tailored to the hydrodata
# and that also contains a small validation step to remove "outdated" values
def _response2dataframe(response,
validate=True,
time_threshold=datetime.timedelta(minutes=30)):
# return pd.read_csv(io.StringIO(resp.text))
data = response.text.strip().split('\n')
columns = data[0].split(',')
data = [i.strip().split(',') for i in data[1:]]
index = [i[0] for i in data]
ix = columns.index('data')
values = [[i[0]] + i[ix].split(';') for i in data]
data = [[j for _, j in enumerate(i) if _ != ix]
for i in data]
# the dataframe holding all non measurement values
columns.pop(ix)
data1 = pd.DataFrame(columns=columns,
data=data,
index=index)
for col in data1.columns:
data1[col] = pd.to_numeric(data1[col], errors='ignore')
def _list2dict(datalist):
return {i: j for i, j in zip(datalist[::2], datalist[1::2])}
values = {i[0]: _list2dict(i[1:])
for i in values}
ix = values.keys()
columns = []
for i in values.items():
vartypes = list(i[1].keys())
for vartype in vartypes:
if vartype in columns:
pass
else:
columns.append(vartype)
data = [[] for i in columns]
for i in ix:
for colno, colname in enumerate(columns):
value = values[i][colname] if colname in values[i].keys() else ''
data[colno].append(value)
data = np.transpose(data)
data2 = pd.DataFrame(columns=columns, data=data, index=ix)
for col in data2.columns:
if col != 'measurementTime':
data2[col] = pd.to_numeric(data2[col], errors='coerce')
data = pd.concat([data1, data2], axis=1)
data['query_time'] = pd.to_datetime(data['query_time'])
data['measurementTime'] = pd.to_datetime(data['measurementTime'])
if validate:
time_threshold = pd.to_timedelta(time_threshold)
good = data['query_time'] <= data['measurementTime'] + time_threshold
# for illustration
bad = data['query_time'] >= data['measurementTime'] + time_threshold
data['measurementAge'] = data['query_time'] - data['measurementTime']
data = data[good]
return dataAdditionally, we can fix the offset that we found (~400 km in latitude) in the geometry by using the following (after importing my conversion functions):
rw, hw = wgs84_to_ch1903(df['latitude'], df['longitude'])
hw = hw + 400000
lat, lon = ch1903_to_wgs84(hw, rw)Now we can see the difference, especially when we plot the dataframe of water temperatures (the labelling is really overkill at this point and just for illustration):
## fix the offset
hw, rw = wgs84_to_ch1903(df['latitude'], df['longitude'], plus=True)
hw = hw + 400000
lat, lon = ch1903_to_wgs84(hw, rw)
# setup the map
import cartopy.crs as ccrs
import cartopy.feature as cf
fig, ax = plt.subplots(dpi=200, figsize=[9, 9], subplot_kw={"projection":ccrs.PlateCarree()})
thevar = 'waterTemperature'
units = {'waterLevel': 'm MSL', 'waterTemperature': '°C', 'discharge': 'm³/s'}
# show the "original" coordinates
s = ax.scatter(df['longitude'], df['latitude'], s=150, edgecolor='k', c=df[thevar], vmin=0, vmax=25, cmap='coolwarm')
# show the fixed coordinates
s = ax.scatter(lon, lat, s=150, edgecolor='k', c=df[thevar], cmap='coolwarm', vmin=0, vmax=25)
# the respective name of the station as label if need be
add_station_label = False
if add_station_label:
for sid, thevalue, lon, lat, station_name in zip(df['station_id'], df[thevar], df['longitude'], df['latitude'], df['station_name']):
if np.isnan(thevalue):
continue
ax.text(lon, lat, station_name.replace(';','\n').replace(' ','\n'), va='center', ha='center', fontsize=3, clip_on=True)
# map decorations
ax.set_xlabel('Longitude (°)')
ax.set_ylabel('Latitude (°)')
plt.colorbar(s, aspect=50, pad=0.1, location='bottom', label=f'{thevar.replace("water","").capitalize()}'+f' ({units[thevar]})')
ax.set_title(f'Hydrodata in Switzerland')
ax.gridlines(draw_labels=True, dms=True, x_inline=False, y_inline=False, lw=0.25, color='k')
# if you want, add a stockimage but it won't look nice at this resolution
#ax.stock_img()
# add coastline and borders to illustrate
ax.coastlines()
ax.add_feature(cf.BORDERS)
# set proper boundaries for illustration
ax.set_ylim(42, 48)
ax.set_xlim(5, 12)If you wish instead to only plot data points that have a valid temperature you can use the condition notnull() on the variable:
# fix the offset
hw, rw = wgs84_to_ch1903(df['latitude'], df['longitude'], plus=True)
hw = hw + 400000
lat, lon = ch1903_to_wgs84(hw, rw)
# setup the map
fig, ax = plt.subplots(dpi=200, figsize=[16, 12], subplot_kw={"projection":ccrs.PlateCarree()})
thevar = 'waterTemperature'
units = {'waterLevel': 'm MSL', 'waterTemperature': '°C', 'discharge': 'm³/s'}
# filter out stations without values for this variable
cond = df[thevar].notnull()
# show the "original" coordinates
s = ax.scatter(df['longitude'][cond], df['latitude'][cond], s=150, edgecolor='k', c=df[thevar][cond], vmin=0, vmax=25, cmap='coolwarm')
# show the fixed coordinates
s = ax.scatter(lon[cond], lat[cond], s=150, edgecolor='k', c=df[thevar][cond], cmap='coolwarm', vmin=0, vmax=25)
# map decorations
ax.set_xlabel('Longitude (°)')
ax.set_ylabel('Latitude (°)')
plt.colorbar(s, aspect=50, pad=0.1, location='bottom', label=f'{thevar.replace("water","").capitalize()}'+f' ({units[thevar]})')
ax.set_title(f'Hydrodata of Switzerland')
ax.gridlines(draw_labels=True, dms=True, x_inline=False, y_inline=False, lw=0.25, color='k')
# if you want, add a stockimage but it won't look nice at this resolution
#ax.stock_img()
# add coastline and borders to illustrate
ax.coastlines()
ax.add_feature(cf.BORDERS)
# set proper boundaries for illustration
ax.set_ylim(45.5, 48)
ax.set_xlim(5.5, 11)Going forward, you could use requests to download the past measurement data that are linked in the dataframe via the query (hydrodata URL) and make an animation; probably xarray would be a better choice for holding the data then though…
Hope this was interesting on how to combine and going forward we’ll look at using openlayers and the JSON response from lindas to make a map.
PS: On another note, while we found an offset last time, there is one station in the dataset that has a wrong longitude as well, the station with id 2270 “Combe des Sarrasins” with latitude of 41.315… and longitude of 0.296… according to its lindas entry but should really be at 47.195… , 6.877…